Salmonid Rickettsial Septicemia (SRS) – Texto en Inglés
What is Salmonid Rickettsial Septicemia?
SALMONID RICKETTSIAL SEPTICEMIA (SRS) is an endemic bacterial disease caused by an intra-cellular bacteria called Piscirickettsia salmonis. This results in lethargy, anorexia and anemia, with the consequent reductions in growth and performance of affected farms. Treatment with antibiotic is used to reduce the effects. A vaccine is available, but it is not completely effective. The disease causes generalized septicemia and produces considerable damages at gastro-intestinal, cutaneous, visceral and nervous levels.
SRS infection results in high mortalities and is in fact the major cause of infectious mortalities in the grow-out phase. SRS is the major reason for the use of antibiotics in the Chilean industry, and its treatment is estimated to cost over $100M per year. Ova with improved resistance to SRS have been available to Benchmark Gentics’ customers in Chile since 2018.
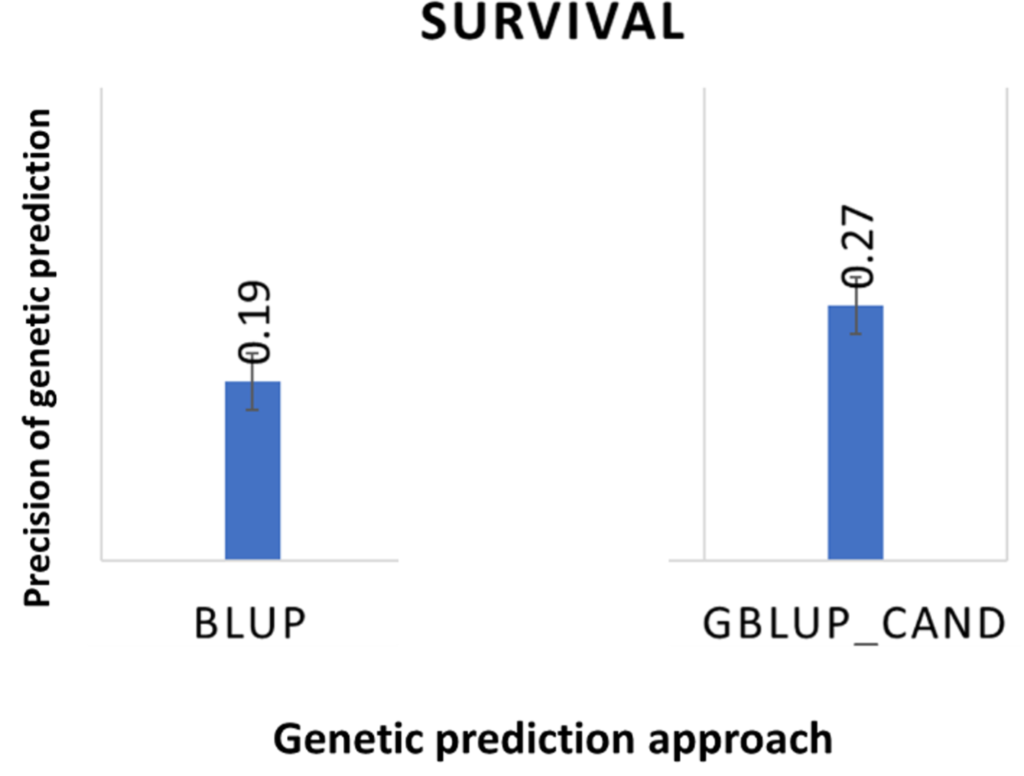
BENCHMARK GENETICS have shown that resistance to SRS is controlled by many genes – polygenic structure – and uses GS to identify broodstock with improved SRS resistance for breeding (Figure 3). Genomic studies indicate that the use of GS increases in a 42% of precision for detecting the best SRS resistant animals (Figure 4).