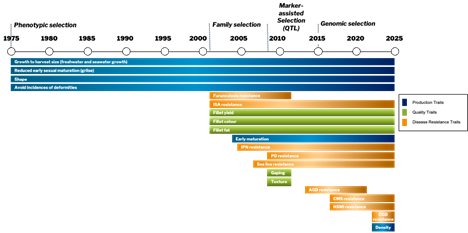
Selection Methods
Moving on to family selection in 2001, new traits came into the program related to harvest quality and disease resistance.
Since 2009, individual selection methods have significantly improved the precision of selection and thereby the genetic gains for important traits.
Our Methods
The Benchmark Genetics in-house breeding programs are all based on family selection.
Why family-based selection?
Before the application of family-based selection, it was only possible to select broodstock as parents of the next generation based on their own performance and for traits that could be directly measured on themselves.
Family-based selection involves the collection of data from siblings of broodstock candidates for traits that cannot be directly measured on the broodstock themselves. Such traits include disease resistance, fillet yield and fillet colour. By comparing families for various traits important in commercial production, we can accurately identify families with specific genetic advantages. Family-based selection is also an effective way of minimising the levels of inbreeding, as we are in control of the pedigrees of each family.
For traits such as growth, where the phenotypes are easily measured on breeding candidates, individuals can be efficiently selected based on their own performance.
However, for the majority of commercially important traits, it is not possible to obtain individual performance of broodstock candidates. Family-based selection, achieved through testing siblings under various conditions, enables the selection of the best families. Still, it is limited in that it does not utilise the within-family variation for traits (i.e. differences in performance between brothers and sisters). It is therefore crucial to apply individual-level selection to increase the precision of selection. This can be achieved by incorporating genomic information into the breeding program. Benchmark Genetics has been using genomic information for individual selection for traits since 2009, and individual selection with genomics has been implemented in all of our in-house breeding programs.
In recent years, new genomic methods have been developed to identify individuals for breeding based on DNA sequence by genotyping. The salmon genome is comprised of approximately 3 billion nucleotide base pairs, all of which can potentially show variation and can affect the performance of an individual for a given trait. These variations are typically called ‘single nucleotide polymorphisms ‘ (or SNPs), of which there are several million in farmed salmon populations. To determine whether an individual is genetically strong or weak for a given trait, it is not necessary to obtain information on every single SNP. Instead, it is possible to identify SNPs that “tag” or track inheritance of neighbouring SNPs. Genotyping of these tag SNPs using an SNP-array (approximately 70K SNPs) enables the accurate estimation of individual performance without genotyping every single SNP.
Genotyping of individuals on this SNP-array enables us to understand the genetics behind each trait and how the genetics is associated with excellent performance, and to screen our populations for the best-performing individuals to use as parents of the next generation without testing them directly.
Depending on the genetic architecture of the trait, selection using genomic information can be split into two categories. If a trait is largely controlled by a single or a few genes, one or a few tag SNPs can be used to track which version (good or bad) each individual has of those genes. The use of a few tag SNPs in selection in this way is known as marker-assisted selection (MAS), and the regions that they track that contain the genes controlling the trait of interest are known as Quantitative Trait Loci (QTL). For traits that are controlled by many thousands of genes located across the genome, information from the full SNP-array is utilised to capture the full genetic effect.
For traits that are controlled by a few quantitative trait loci (QTL) with large effects, it is possible to select only for these QTL using marker-assisted selection.
In Atlantic salmon, a significant QTL for resistance to IPN virus was identified in 2007. This single QTL accounts for over 80% of the genetic variation in resistance, and, therefore, selecting only on this QTL has achieved significant reductions in IPN-related mortality since the inclusion of MAS for IPN resistance in 2009. SNP markers tagging this QTL are used to identify and breed from genetically resistant individuals in the nucleus, as well as for commercial egg production, thereby reducing the number and severity of IPN outbreaks.
The IPN QTL has been one of the most successful examples of using genetics to help control an infectious disease in animal farming. Though researchers continue to identify significant QTL for other traits, these tend to be of small effect. For most traits, the majority of variation is explained by many genes, each of which has a small effect, but together they explain the performance of an individual. In this case, Genomic Selection is more effective.
Marker-assisted selection for larger effect QTL is a very effective method for traits where a major gene accounts for the majority of genetic variation. However, for most traits, genetic variation is controlled by a large number of genes, each with a small effect. For these traits, Genomic Selection (GS) is more effective in identifying individuals for breeding.
Traits selected using genomic selection include (but are not limited to) growth, delayed sexual maturation, disease resistance and flesh quality. Our breeding programs for Atlantic salmon are internationally recognized for the quality of our products for the traits where we apply GS.